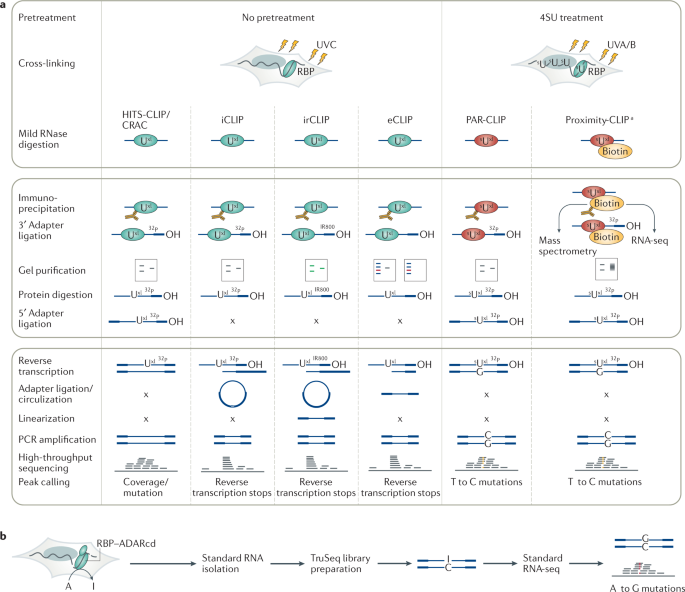
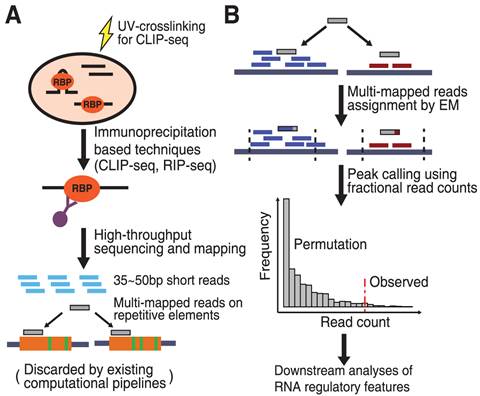
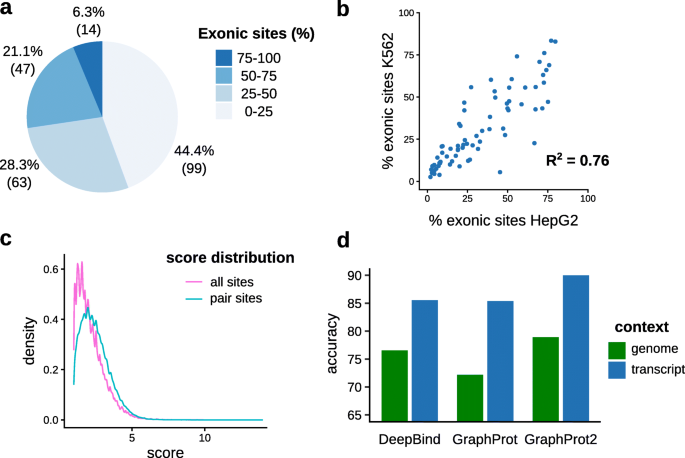
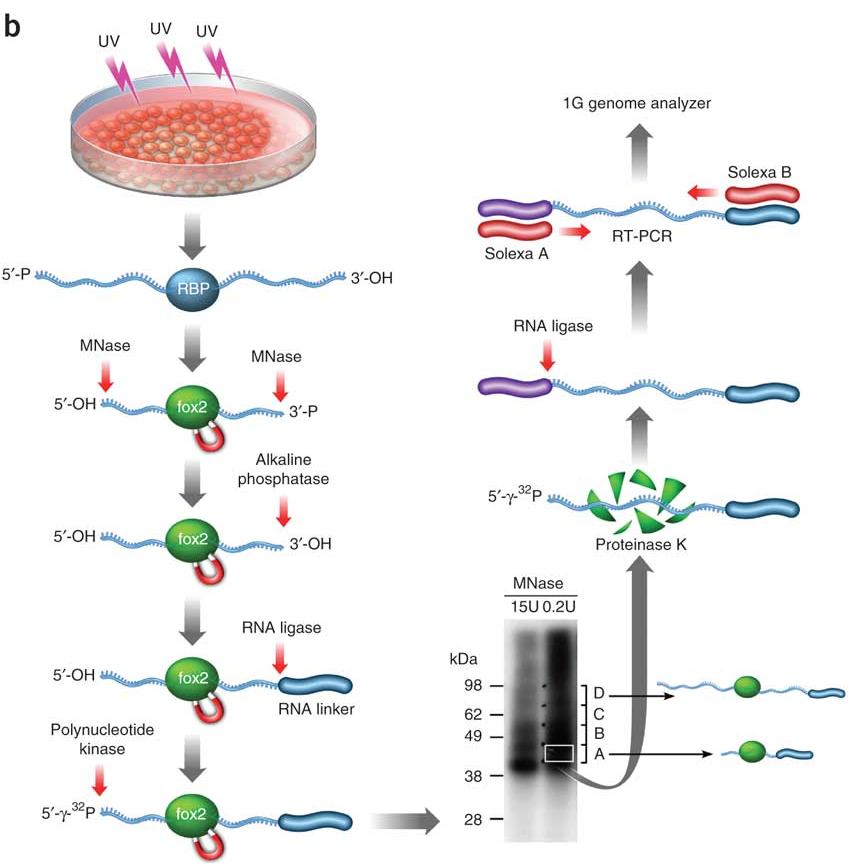
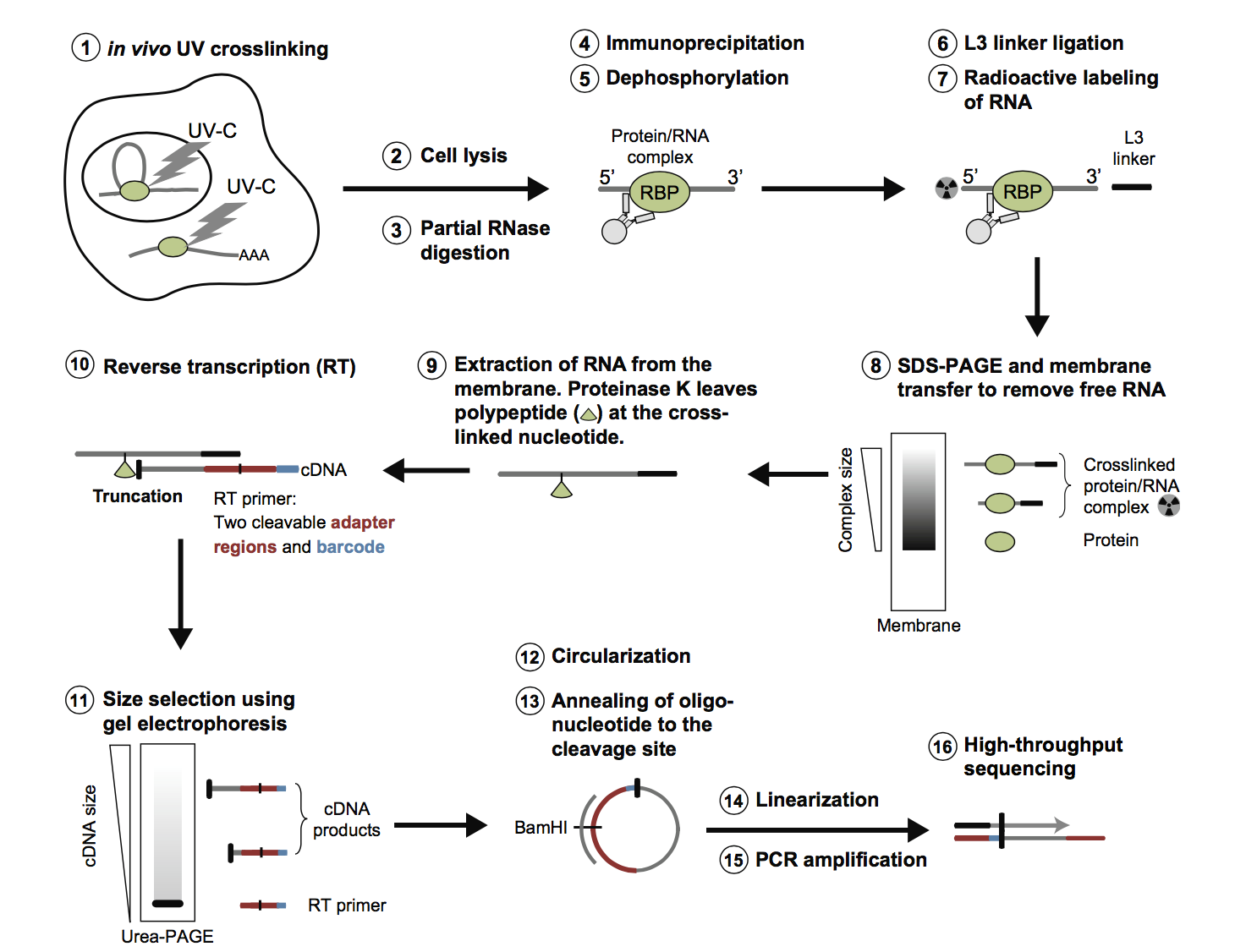
Summary of the analysis software, pipelines and databases for CLIP-Seq... | Download Scientific Diagram

omniCLIP: probabilistic identification of protein-RNA interactions from CLIP -seq data | RNA-Seq Blog

Main steps of the bioinformatics workflow to analyze CLIP-seq data with... | Download Scientific Diagram
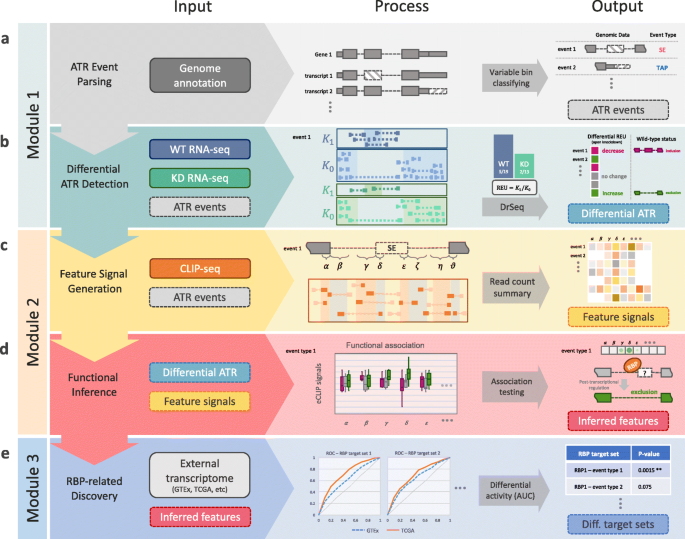
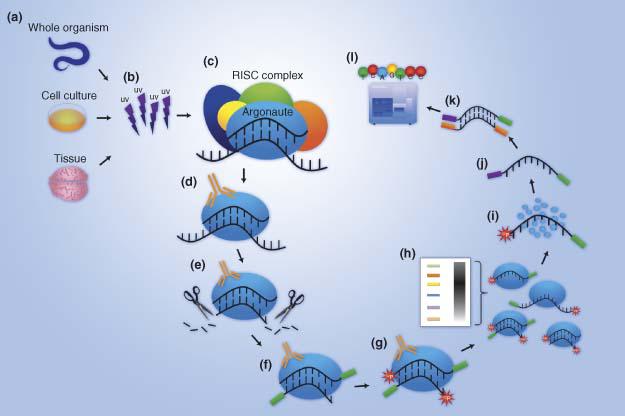
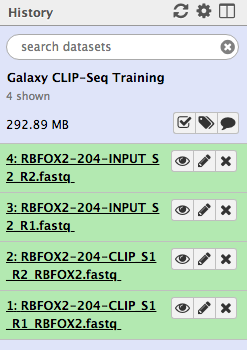
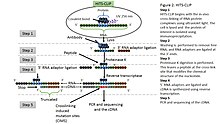
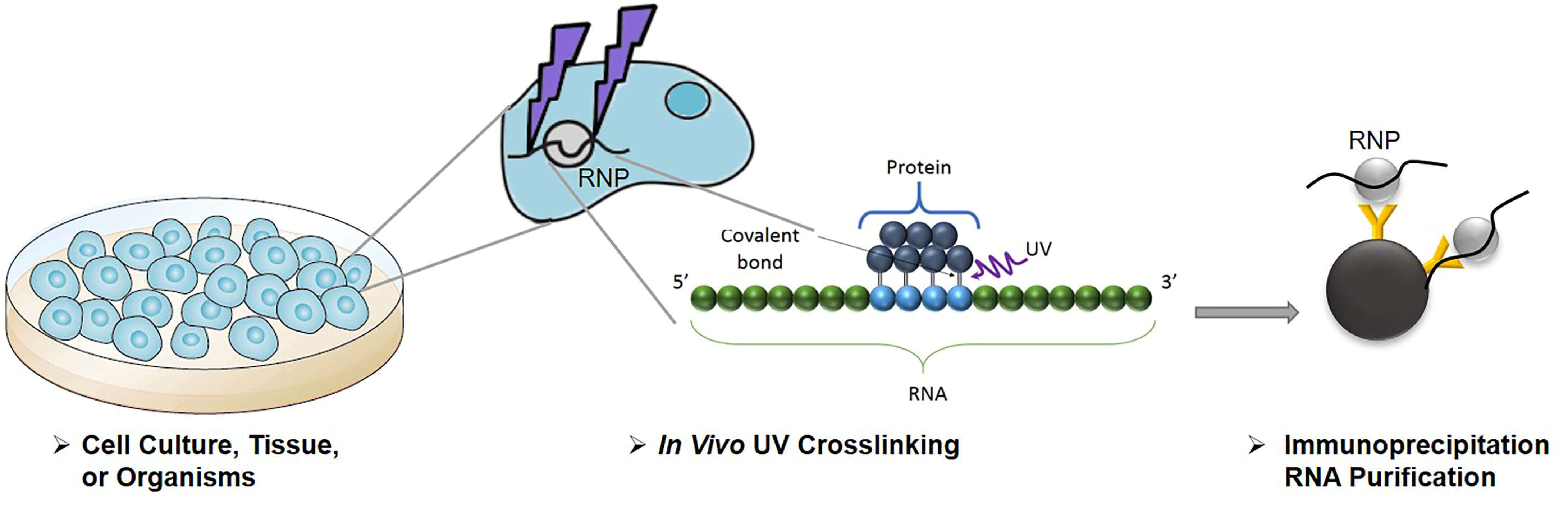

![PDF] CLIPSeqTools—a novel bioinformatics CLIP-seq analysis suite | Semantic Scholar PDF] CLIPSeqTools—a novel bioinformatics CLIP-seq analysis suite | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/6bbd241680ec0f3d6a86dec738a6b864ea5048c3/6-Figure3-1.png)