Frontiers | Emerging Perspectives of RNA N6-methyladenosine (m6A) Modification on Immunity and Autoimmune Diseases

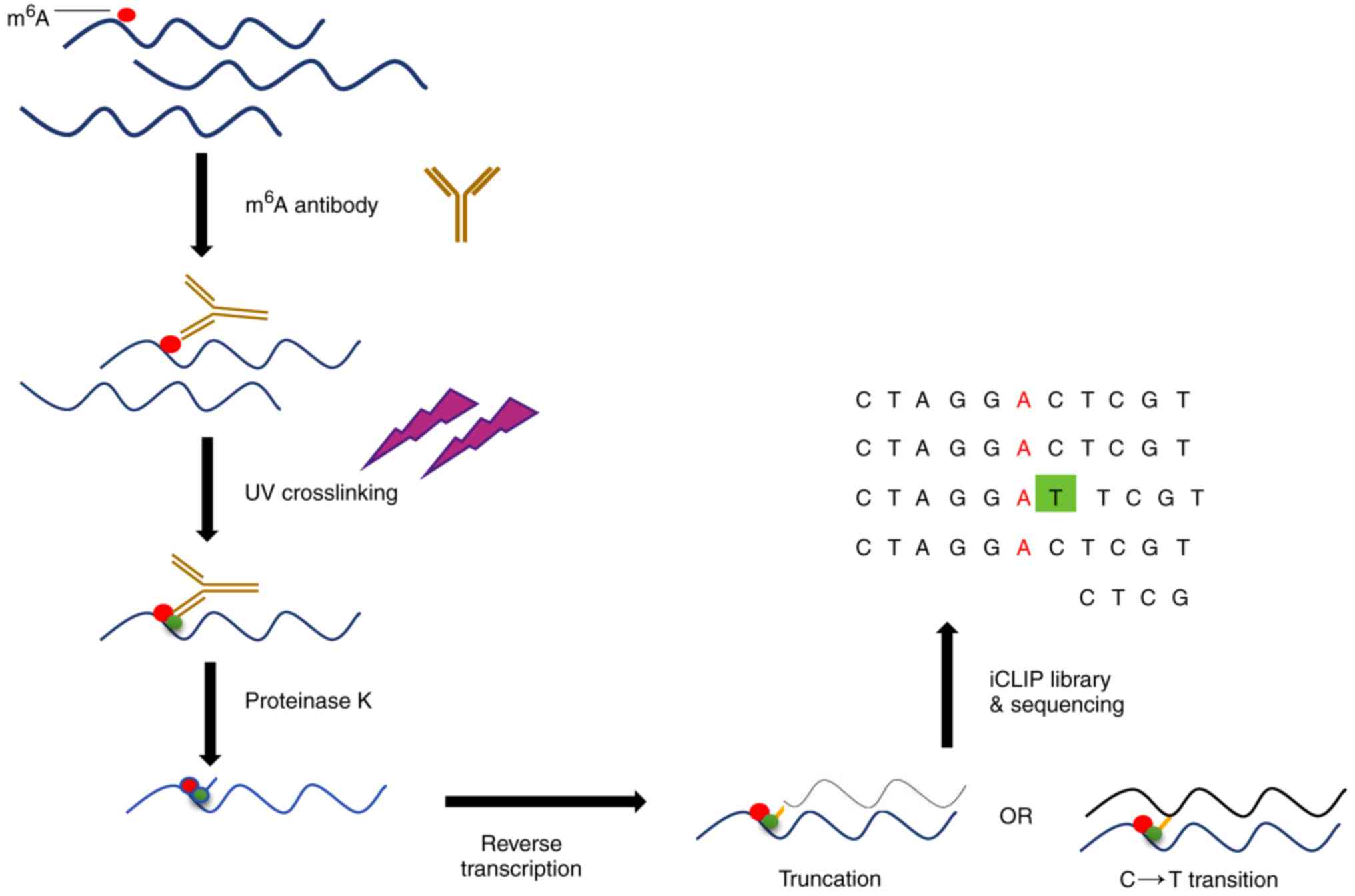
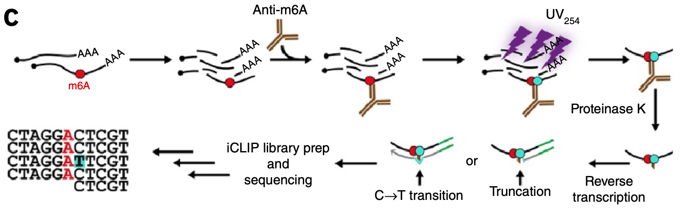
High‐Resolution N6‐Methyladenosine (m6A) Map Using Photo‐Crosslinking‐Assisted m6A Sequencing - Chen - 2015 - Angewandte Chemie International Edition - Wiley Online Library

Deep and accurate detection of m6A RNA modifications using miCLIP2 and m6Aboost machine learning | bioRxiv
![PDF] High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. | Semantic Scholar PDF] High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/e1a071ac484a9f518fb5a1fe468ffeaa06250139/3-Figure2-1.png)
PDF] High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. | Semantic Scholar

Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine | Nature Chemical Biology
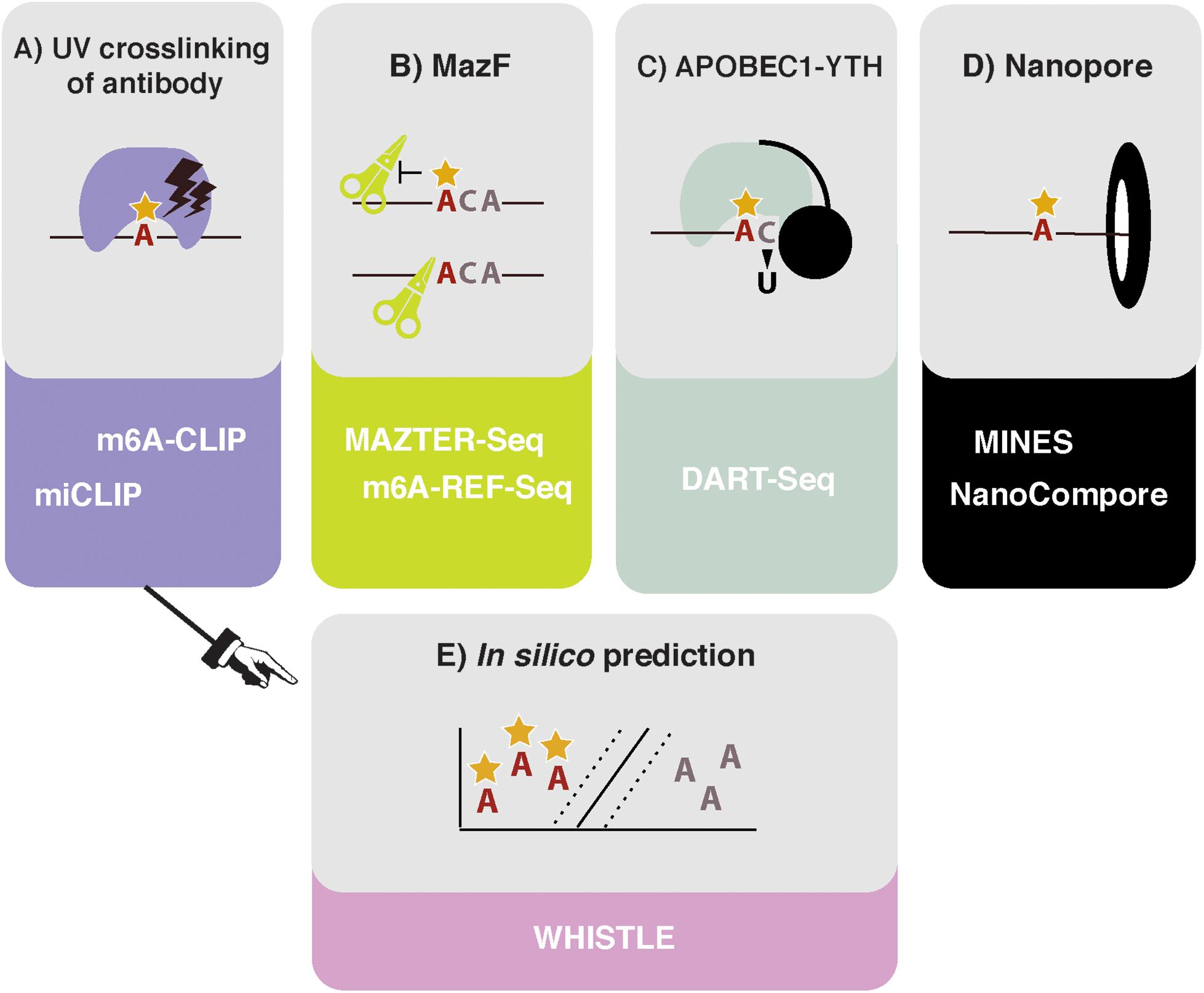
Frontiers | How Do You Identify m6 A Methylation in Transcriptomes at High Resolution? A Comparison of Recent Datasets

Acute depletion of METTL3 identifies a role for N6-methyladenosine in alternative intron/exon inclusion in the nascent transcriptome | bioRxiv

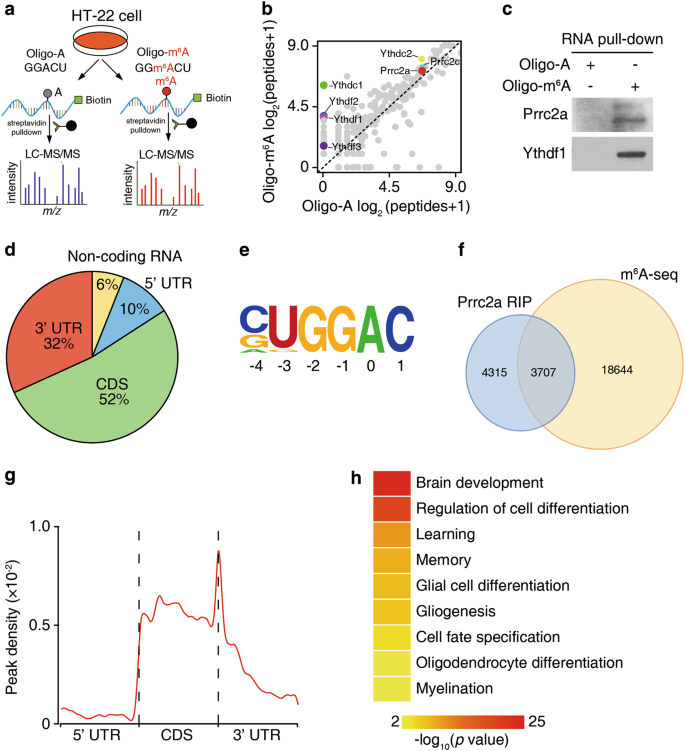
RNA m6A modification orchestrates a LINE-1–host interaction that facilitates retrotransposition and contributes to long gene vulnerability | Cell Research

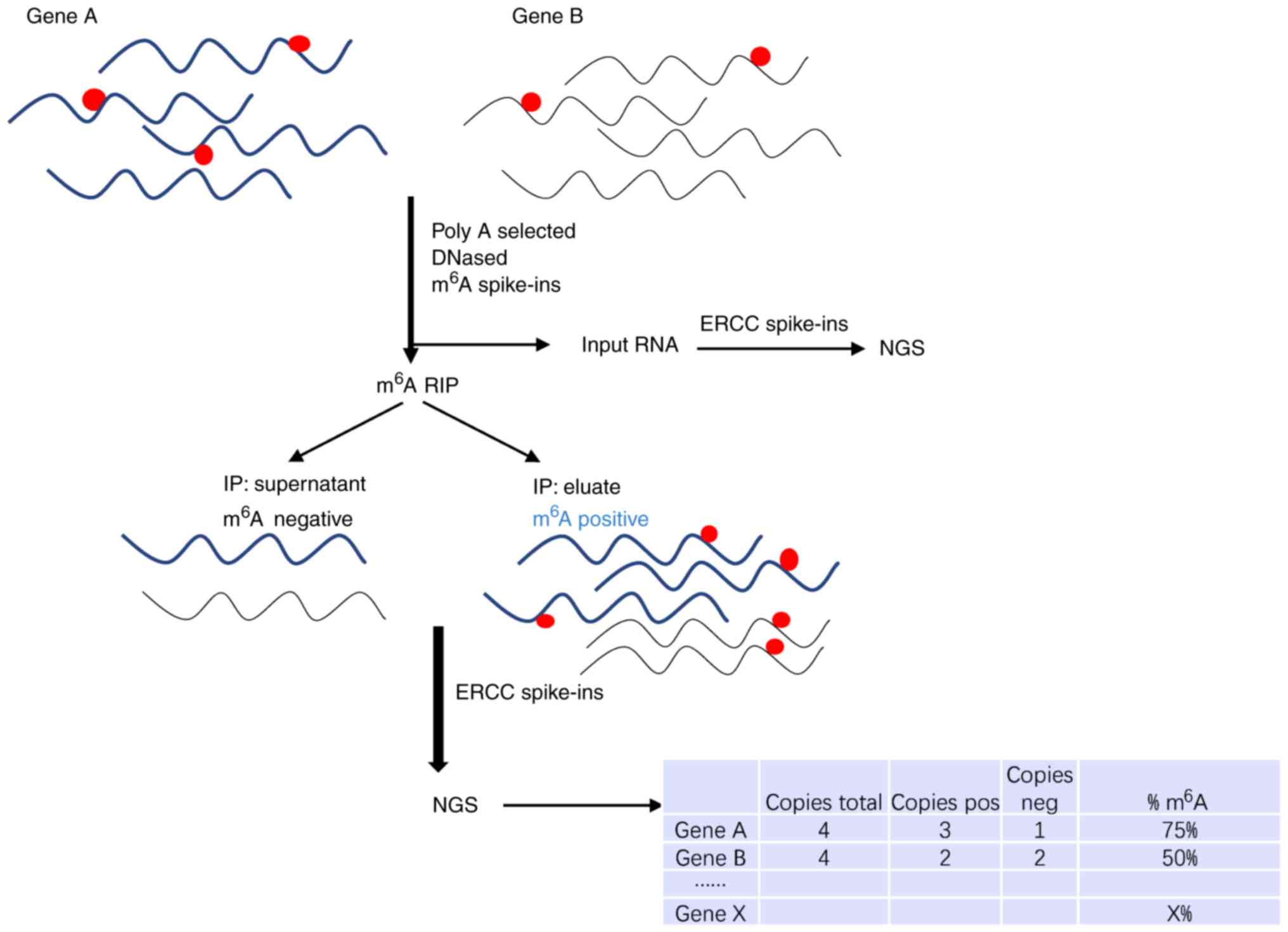
Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing | Nature Protocols
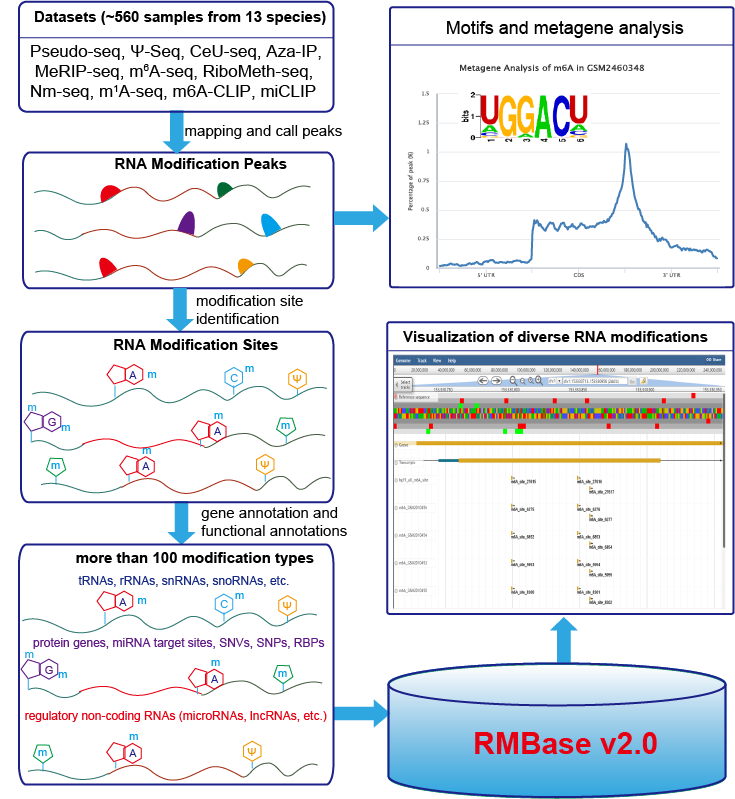
RNA Modification Database, RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data (Pseudo-seq, Ψ-seq, CeU-seq, Aza-IP, MeRIP-seq, m6A-seq, RiboMeth-seq, m1A-seq).
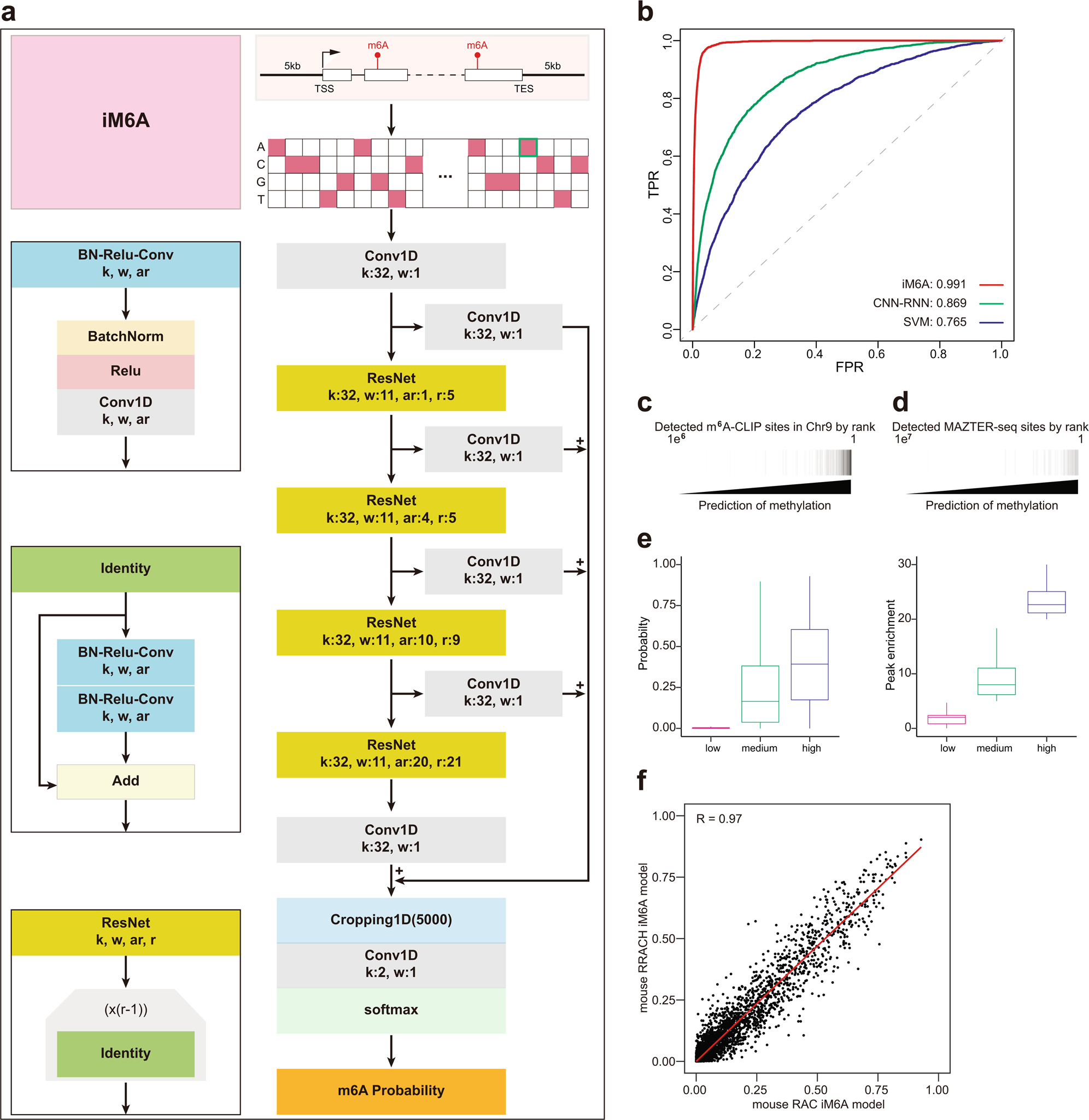
Deep learning modeling m6A deposition reveals the importance of downstream cis-element sequences | Nature Communications

Down-Regulation of m6A mRNA Methylation Is Involved in Dopaminergic Neuronal Death | ACS Chemical Neuroscience